How would Alan Turing develop biology?
June 23, 2012 29 Comments
Alan Turing was born 100 years ago, today: June 23rd, 1912. He was a pioneer of computing, cryptography, artificial intelligence, and biology. His most influential work was launching computer science by the definition of computable, introduction of Turing-machine, and solution of the Entscheidungsproblem (Turing, 1936). He served his King and Country in WW2 as the leader of Hut 8 in the Government Code and Cypher School at Bletchley Park. With his genius the British were able to build a semi-automated system for cracking the Enigma machine used for German encryption. After the war he foresaw the connectionist-movement of Cognitive Science by developing his B-type neural network in 1948. He launched the field of artificial intelligence with Computing machinery and intelligence (1950), introducing the still discussed Turing test. In 1952 he published his most cited work: The Chemical Basis of Morphogenesis spurring the development of mathematical biology. Unfortunately, Turing did not leave to see his impact on biology.
In 1952, homosexuality was illegal in the United Kingdom and Turing’s sexual orientation was criminally prosecuted. As an alternative to prison he accepted chemical castration (treatement with female hormones). On June 8th, 1954, just two weeks shy of his 42nd birthday, Alan Turing was found dead in his apartment. He died of cyanide poisoning, and an inquire ruled the death a suicide. A visionary pioneer was taken and we can only wonder: how would Alan Turing develop biology?
In The Chemical Basis of Morphogenesis (1952) Turing asked: how does a spherically symmetric embryo develop into a non-spherically symmetric organism under the action of symmetry-preserving chemical diffusion of morphogens? Morphogens are abstract particles that Turing defined; they can stand in place for any molecules relevant to developmental biology. The key insight that Turing made is that very small stochastic fluctuations in the chemical distribution can be amplified by diffusion to produce stable patterns that break the spherical symmetry. These asymmetric patters are stable and can be time-independent (except a slow increase in intensity), although with three or more morphogens there is also the potential for time-varying patterns.
The beauty of Turing’s work was in its abstraction and simplicity. He modeled the question generally via Chemical diffusion equations and instantiated his model by considering specific arrangements of cells like a discrete cycle, and a continuous ring of tissue. He proved results that were general and qualitative in nature. On more complicated models he also encouraged a numeric quantitative approach to be carried out on the computer he helped develop. It is these rigorous qualitative statements that have become the bread-and-butter of theoretical computer science (TCS).
For me, rigorous qualitative statements (valid for various constants and parameters) instead of quantitative statements based on specific (in some fields: potentially impossible to measure) constant and parameters is one of the two things that sets TCS apart from theoretical physics. The other key feature is that TCS deals with discrete objects of arbitrarily large size, while (classical) physics assumes that the relevant behavior can be approximated by continuous variables. The differential equation approach of physics can provide useful approximations such as replicator dynamics (example applications: perception-deception and cost-of-agency), I think it is fundamentally limited. Differential equations should only be used for intuition in fields like theoretical biology. Although Turing did not get a chance to pursue this avenue, I think that he would have pushed biology into the direction of using more discrete models.
Shnerb et al. (2000) make a good point for the importance of discrete models. The model is of spatial diffusion with two populations: catalyst A that never expires and a population B — agents of which expire at a constant rate and use A to catalyze reproduction. The authors use the standard mean-field diffusion approach to look at the parameter range where the abundance of A is not high enough to counteract the death rate of B agents. The macro-dynamic differential equation approach predicts extinction of the B-population at an exponential rate. However, when the model is simulated at micro-dynamic level with discrete agents, then there is no extinction. Clumps of B agents form and follow individual A agents as they follow Brownian motion through the population. The result is an abundance of life (B agents) at the macro-scale in contrast to the continuous approximation. This is beautifully summarized by Shnerb et al. (2000) in the figure below.
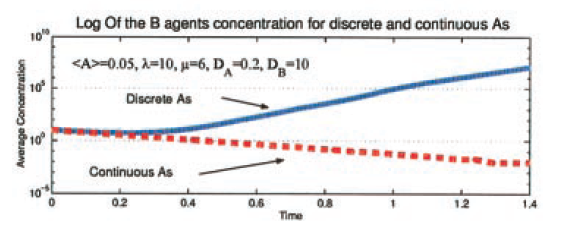
Figure 1 from Shnerb et al. (2000) showing Log of B agent concentration versus time for discrete (solid blue line) and continues (dotted red line) models.
Like Turing’s (1952) approach, the discrete model also shows clumping and symmetry breaking. However, the requirements are not as demanding as what Turing developed. Thus, it is natural to expect that Turing would have found similar models if he continued his work on morphogenesis. This is made more likely by Turing’s exploration of discrete computer models of Artificial Life prior to his death. I think that he would have developed biology by promoting the approach of theoretical computer science: simple abstract models that lead to rigorous qualitative results about discrete structures; Alan Turing would view biology through algorithmic lenses. Since he is no longer with us, I hope that myself and others can carry on his vision.
References
Shnerb NM, Louzoun Y, Bettelheim E, & Solomon S (2000). The importance of being discrete: Life always wins on the surface. Proceedings of the National Academy of Sciences of the United States of America, 97 (19), 10322-4 PMID: 10962027
Turing, A. M. (1936). On Computable Numbers, with an Application to the Entscheidungsproblem. Proceedings of the London Mathematical Society 2(42): 230–65.
Turing, A. M. (1950) Computing Machinery and Intelligence. Mind.
Turing, A. M. (1952). The Chemical Basis of Morphogenesis. Philosophical Transactions of the Royal Society of London 237 (641): 37–72. DOI:10.1098/rstb.1952.0012


I think it would be nice if you could expand on and explain why there is a different prediction/outcome with the different macro/continuous vs micro/discrete approaches. I don’t have much physics background and do not know what standard mean-field diffusion is, but I’m curious how these contrasting results come about.
They modeled their process analytically as diffusion on a finite lattice. This has a standard description in terms of differential equations. The reason I say they used a mean-field approach to diffusion is because they assumed a long time scale on which the A-types where at equilibrium in the population. This allows them to reduce relatively complicated DEs to very simple ordinary differential equations and solve the model. The function that satisfies their differential equation is exponential decay; hence their analytic solution.
For the micro approach they ran simulations and then a more involved renormalization group (RG) treatment of their simulations modeled as a type of random walk. The second treatment is presented elsewhere and not essential to their results. This lets them say how dimension of the lattice effects the results, and provides a more thorough analysis. However, I have not tracked down the ‘elsewhere’ to look at the RG approach.
Usually RGs get very complicated. I do not have a strong grasp of them, but it is a standard tool in statistical mechanics where it was developed to handle cases where the mean-field approach fails. If my memory serves me correctly, it was first used to deal with the square lattice Ising model, so it is natural that the approach pops up when trying to analyze the sort of lattice model they looked at in their paper.
you wouldn’t happen to have the matlab files for the continuum and discret models of the figures you showed above would you? its one of the projects i have to do for an assignment and i just couldn’t get this results that Shnerb showed
Pingback: How would Alan Turing develop biology? | Gavagai | Scoop.it
Pingback: Bifurcation of cooperation and inviscid ethnocentrism « Theory, Evolution, and Games Group
Pingback: How would Alan Turing develop biology? | Modern Biology: of computers and men | Scoop.it
Pingback: EGT Reading Group 21 – 30 « Theory, Evolution, and Games Group
Pingback: How would Alan Turing develop biology? | Gentlemachines | Scoop.it
Pingback: How would Alan Turing develop biology? | this curious life | Scoop.it
Pingback: Theorists as connectors: from Poincaré to mathematical medicine « Theory, Evolution, and Games Group
Pingback: Some stats on the first 50 posts « Theory, Evolution, and Games Group
It seems to me, a relative mathematical babe-in-the-woods, that Turing’s work here has another significant implication: abiogenesis. Using your description ” He modeled the question generally via Chemical diffusion equations and instantiated his model by considering specific arrangements of cells like a discrete cycle, and a continuous ring of tissue. He proved results that were general and qualitative in nature. Another description I saw was:” the self-assembly of single-celled organisms having no neonatal precursor” . When considering the simplest forms of life, small enclosed blobs of RNA essentially, it would seem that this would meet Turing’s criteria, though he may not specifically have intended his original work to be related directly to abiogenesis. Thoughts?
Thanks for the comment! Sorry that is has taken me so long to get back to you.
I don’t know where you heard the description ”the self-assembly of single-celled organisms having no neonatal precursor”. Morphogenesis is the study of how a spherically symmetric egg (which obviously has a precursor) manages to disrupt parts of its symmetry in a systematic way while developing under symmetric laws. The question has nothing to do (at least in the way Turing studied it) with evolution, or abiogenesis. It is mostly a question of biomechanics. The only thing is common is the root “genesis”, but in the first case it means gaining a distinct morphology when one did not exist, while in the second it means becoming biological from a non-biological ancestor. I doubt there are that many direct connections or too much overlap in tools.
Some of Turing’s work (especially his late unpublished work on artifical life simulations) could be loosely linked to abiogenesis. However, since the work was not published, I doubt it would have had a chance to produce much of an impact (much like Turing’s role in the first neural networks is virtually unknown to both the public and scientists who works in the subject).
For further reference see is my list of ways Turing contributed to science by viewing it through the algorithmic lens.
Pingback: Games, culture, and the Turing test (Part I) | Theory, Evolution, and Games Group
Pingback: Natural algorithms and the sciences | Theory, Evolution, and Games Group
Pingback: Algorithmic view of historicity and separation of scales in biology | Theory, Evolution, and Games Group
Pingback: Microscopic computing in cells and with self-assembling DNA tiles | Theory, Evolution, and Games Group
Pingback: Computational complexity of evolutionary equilibria | Theory, Evolution, and Games Group
Pingback: Semi-smooth fitness landscapes and the simplex algorithm | Theory, Evolution, and Games Group
Pingback: Simplifying models of stem-cell dynamics in chronic myeloid leukemia | Theory, Evolution, and Games Group
Pingback: Phenotypic plasticity, learning, and evolution | Theory, Evolution, and Games Group
Pingback: Evolution is a special kind of (machine) learning | Theory, Evolution, and Games Group
Pingback: Ecology of cancer: mimicry, eco-engineers, morphostats, and nutrition | Theory, Evolution, and Games Group
Pingback: Stem cells, branching processes and stochasticity in cancer | Theory, Evolution, and Games Group
Pingback: Space and stochasticity in evolutionary games | Theory, Evolution, and Games Group
Pingback: Choosing units of size for populations of cells | Theory, Evolution, and Games Group
Pingback: Multiple realizability of replicator dynamics | Theory, Evolution, and Games Group
Pingback: Algorithmic lens as Alan Turing’s wider impact | Theory, Evolution, and Games Group
Pingback: Mathtimidation by analytic solution vs curse of computing by simulation | Theory, Evolution, and Games Group