October 22, 2012
by Artem Kaznatcheev
Last week, we saw how to deal with two-strategy games in finite inviscid populations. Unfortunately, two strategy games are not adequate to model many of the interactions we might be interested in. In particular, we cannot use Antal et al. (2009a) to look at bifurcations and finitary/stochastic effects in tag-based models of ethnocentrism, at least not without some subtle tricks. In this post we are going to look at a more general approach for any  -strategy game developed by Antal et al. (2009b). We will apply these tools to look at a new problem for the paper, but an old problem for the blog: ethnocentrism.
-strategy game developed by Antal et al. (2009b). We will apply these tools to look at a new problem for the paper, but an old problem for the blog: ethnocentrism.
Antal et al. (2009b) consider a large but finite population of size 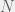 and a game
and a game  with
with  strategies. For update rule, they focus on the frequency dependent Moran process, although their results also hold for Wright-Fisher, and pairwise comparison (Fermi rule). Unlike the previous results for two-strategy games, the present work is applicable only in the limit of weak selection. Mutations are assumed to be uniform with probability
strategies. For update rule, they focus on the frequency dependent Moran process, although their results also hold for Wright-Fisher, and pairwise comparison (Fermi rule). Unlike the previous results for two-strategy games, the present work is applicable only in the limit of weak selection. Mutations are assumed to be uniform with probability  : a mutant is any one of the
: a mutant is any one of the  strategies with equal probabilities. However, in section 4.2, the authors provide a cute argument for approximating non-uniform but parent-independent mutation rates by repeating strategies in proportion to the likelihood of mutation into them. Much like the two-strategy case, the authors derive separate equations for low and high mutation rates, and then provide a way to interpolate between the two extremes for intermediate mutation rates.
strategies with equal probabilities. However, in section 4.2, the authors provide a cute argument for approximating non-uniform but parent-independent mutation rates by repeating strategies in proportion to the likelihood of mutation into them. Much like the two-strategy case, the authors derive separate equations for low and high mutation rates, and then provide a way to interpolate between the two extremes for intermediate mutation rates.
The mathematics proceeds through a perturbation analysis. The basic idea is to solve for the distribution of strategies in the drift (no selection) model, and then to gradually dial up the selection to perturb the distribution slightly into the weak selection regime. The authors use this to arrive at the following to conditions for low, and high mutation for strategy 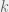 to be favored:
to be favored:
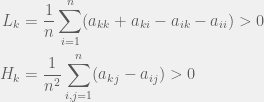
There is an intuitive story to help understand these two conditions. In the case of low mutation, the population is usually just two strategies until one of them fixes. So for  the terms that matter are the pairwise interactions between the strategies, and since the most decisive (and entropy rich) case is near the 50-50 split in the two strategies, self interactions (
the terms that matter are the pairwise interactions between the strategies, and since the most decisive (and entropy rich) case is near the 50-50 split in the two strategies, self interactions ( ) and other-strategy interactions (
) and other-strategy interactions ( ) happen with the same frequency. I think this is where the discrepancy with Antal et. al (2009a) that we will see later sneaks in. A correction factor of
) happen with the same frequency. I think this is where the discrepancy with Antal et. al (2009a) that we will see later sneaks in. A correction factor of  should be added in front of the the self-interaction terms, but I digress.
should be added in front of the the self-interaction terms, but I digress.
For the high mutation case, all the strategies are present in the population with about the same frequency at the same time. We need to look at the transitions from this population to get our first order terms. In that case, the focal individual’s fitness is  (since all opponents are equally likely; once again, I believe a correction term is in order), and the average fitness is
(since all opponents are equally likely; once again, I believe a correction term is in order), and the average fitness is  . The difference of these two terms produces
. The difference of these two terms produces  .
.
In order to interpolate, we have the following condition for strategy 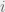 to be more common than
to be more common than  for intermediate mutation rates
for intermediate mutation rates  :
:
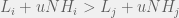
How does this disagree with the two-strategy results of Antal et al. (2009a)? The present paper reproduces the condition of risk-dominance, with C dominating D if 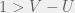 , but does not produce the small
, but does not produce the small 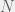 correction of
correction of  . This would be mitigated with the observations I made earlier, but the approach of the perturbation analysis would have to be modified carefully.
. This would be mitigated with the observations I made earlier, but the approach of the perturbation analysis would have to be modified carefully.
The perturbation method can be extended to mixed strategies, as is done by Tarnita et al. (2009). In that case, we just replace the summation by integrals, to get:
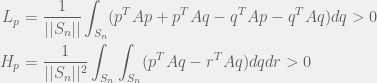
Where 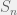 is the n-vertex simplex with volume
is the n-vertex simplex with volume  . It is nice to know that the results generalize to mixed strategies, but not as important tool as the pure strategy variant. I will concentrate on pure strategies, although mixed might be good to revisit to study evolution of agency.
. It is nice to know that the results generalize to mixed strategies, but not as important tool as the pure strategy variant. I will concentrate on pure strategies, although mixed might be good to revisit to study evolution of agency.
Antal et al. (2009b) showcase their method with three case studies: (i) cooperators, defectors, and loners, (ii) reversing the rank of strategies by mutation, and (iii) cooperators, defectors, and tit-for-tat. The first is the most interesting for me, since it shows how adding an irrelevant strategy, can reverse the dominance of the other two. I will present their example in a more general context for all cooperate-defect games. We will introduce an irrelevant strategy L, where irrelevance means that both C and D get the same payoff 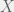 from interacting with L and L gets
from interacting with L and L gets 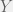 from them. The self interaction payoff for L can differ, and we can set it to
from them. The self interaction payoff for L can differ, and we can set it to  :
:
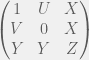
The authors consider the particular case of  and
and 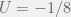 , and
, and  . We can apply the general results to get (for small mutations):
. We can apply the general results to get (for small mutations):
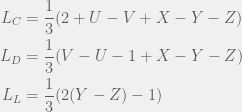
We can look at at the condition for C to dominate D for small mutations ( ) to get
) to get  . If we had used
. If we had used 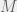 different irrelevant strategies (or just dialed up the proportion of mutations to an irrelevant strategy) then
different irrelevant strategies (or just dialed up the proportion of mutations to an irrelevant strategy) then  would be replaced by
would be replaced by  . This creates a new strip of cooperation which reaches into the Prisoner’s dilemma region (drawn in blue):
. This creates a new strip of cooperation which reaches into the Prisoner’s dilemma region (drawn in blue):

When we switch to large mutations, the region disappears and we recover the standard 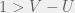 rule. Note that this example means that, for this analysis, we cannot ignore competing strategies even if they are strictly dominated.
rule. Note that this example means that, for this analysis, we cannot ignore competing strategies even if they are strictly dominated.
Ethnocentrism in tag-based models
We will consider a strategy space of  tags, with an agent of each tag being either a humanitarian (cooperate with all), ethnocentric (cooperate with same-tag), traitorous (cooperate with oot-tags), or selfish. For the game, we will look at the usual cost-benefit representation of Prisoner’s dilemma. Note that the
tags, with an agent of each tag being either a humanitarian (cooperate with all), ethnocentric (cooperate with same-tag), traitorous (cooperate with oot-tags), or selfish. For the game, we will look at the usual cost-benefit representation of Prisoner’s dilemma. Note that the  and
and 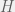 values will be the withing a single strategy of different tags, so we only need to compute four of each:
values will be the withing a single strategy of different tags, so we only need to compute four of each:
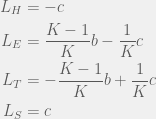
This divides the dynamics into two regions, when 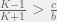 then
then  , otherwise we have
, otherwise we have  . In other words, for large enough
. In other words, for large enough  or small enough
or small enough  , ethnocentrism can be the dominant strategy in the population. This condition is in perfect agreement with the Traulsen & Nowak (2007) results we saw earlier. Although in that case, there were no H or T agents. If we remove H & T from the current analysis, we will still get the same condition for ethnocentric dominance even though we will calculate different
, ethnocentrism can be the dominant strategy in the population. This condition is in perfect agreement with the Traulsen & Nowak (2007) results we saw earlier. Although in that case, there were no H or T agents. If we remove H & T from the current analysis, we will still get the same condition for ethnocentric dominance even though we will calculate different  values.
values.
For large mutations, the advantage of ethnocentrics disappears completely, and we get:
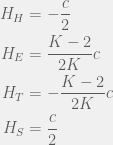
Which for  results in the ordering
results in the ordering 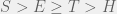 . So if we have mutations that change tag and strategy together (as they do in this case) then higher mutation rates disadvantage the population, and if we let
. So if we have mutations that change tag and strategy together (as they do in this case) then higher mutation rates disadvantage the population, and if we let  be the expected number of mutants per generation, then we can see that ethnocentric cooperation is possible only if
be the expected number of mutants per generation, then we can see that ethnocentric cooperation is possible only if  or rewritten as
or rewritten as 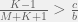 .
.
References
Antal, T., Nowak, M.A., & Traulsen, A. (2009a). Strategy abundance in games for arbitrary mutation rates Journal of Theoretical Biology, 257 (2), 340-344.
Antal T, Traulsen A, Ohtsuki H, Tarnita CE, & Nowak MA (2009b). Mutation-selection equilibrium in games with multiple strategies. Journal of Theoretical Biology, 258 (4), 614-22 PMID: 19248791
Tarnita, C.E., Antal, T., Nowak, M.A. (2009) Mutation-selection equilibrium in games with mixed strategies. Journal of Theoretical Biology 26(1): 50-57.
Traulsen A, & Nowak MA (2007). Chromodynamics of cooperation in finite populations. PLoS One, 2 (3).
(Bianconi et al., 2013).
 Both 206 and
Both 206 and are just numbers, but to a modeler they suggest a very important distinction over which tools we should use. Suppose that my bones and cells randomly popped in and out of existence without about equal probability (thus keeping the average number constant). In that case I wouldn’t expect to see exactly 206 bones, or exactly 37200000000000 cells; if I do a quick back-of-the-envelope calculation then I’d expect to see somewhere between 191 and 220 bones, and between 37199994000000 and 37200006000000. Unsurprisingly, the variance in the number of bones is only around 29 bones, while the number of cells varies by around 12 million. However, in terms of the percentage, I have 14% variance for the bones and only 0.00003% variance in the cell count. This means that in terms of dynamic models, I would be perfectly happy to model the cell population by their average, since the stochastic fluctuations are irrelevant, but — for the bones — a 14% fluctuation is noticeable so I would need to worry about the individual bones (and we do; we even give them names!) instead of approximating them by an average. The small numbers would be a case of when results can depend heavily on if one picks a discrete or continuous model.




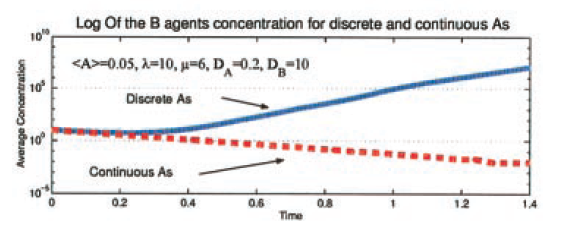

Space and stochasticity in evolutionary games
January 28, 2015 by Artem Kaznatcheev 20 Comments
Two of my goals for TheEGG this year are to expand the line up of contributors and to extend the blog into a publicly accessible venue for active debate about preliminary, in-progress, and published projects; a window into the everyday challenges and miracles of research. Toward the first goal, we have new contributions from Jill Gallaher late last year and Alexander Yartsev this year with more posts taking shape as drafts from Alex, Marcel Montrey, Dan Nichol, Sergio Graziosi, Milo Johnson, and others. For the second goal, we have an exciting debate unfolding that was started when my overview of Archetti (2013,2014) prompted an objection from Philip Gerlee in the comments and Philipp Altrock on twitter. Subsequently, Philip and Philipp combined their objections into a guest post that begat an exciting comment thread with thoughtful discussion between David Basanta, Robert Vander Velde, Marc Harper, and Philip. Last Thursday, I wrote about how my on-going project with Robert, David, and Jacob Scott is expanding on Archetti’s work and was surprised to learn that Philip has responded on twitter with the same criticism as before. I was a little flabbergast by this because I thought that I had already addressed Philip’s critique in my original comment response and that he was reiterating the same exact text in his guest post simply for completeness and record, not because he thought it was still a fool-proof objection.
My biggest concern now is the possibility that Philip and I are talking past each other instead of engaging in a mutually beneficial dialogue. As such, I will use this post to restate (my understand of the relevant parts of) Philip and Philipp’s argument and extend it further, providing a massive bibliography for readers interested in delving deeper into this. In a future post, I will offer a more careful statement of my response. Hopefully Philip or other readers will clarify any misunderstandings or misrepresentations in my summary or extension. Since this discussion started in the context of mathematical oncology, I will occasionally reference cancer, but the primary point at issue is one that should be of interest to all evolutionary game theorists (maybe even most mathematical modelers): the model complexity versus simplicity tension that arises from the stochastic to deterministic transition and the discrete to continuous transition.
Read more of this post
Filed under Commentary, Models, Preliminary, Reviews, Technical Tagged with current events, discrete vs continuous, mathematical oncology, metamodeling, replicator dynamics, spatial structure