Microscopic computing in cells and with self-assembling DNA tiles
May 29, 2013 9 Comments
One of the three goals of natural algorithms is to implement computers in non-electronic media. In cases like quantum computing, the goal is to achieve a qualitatively different form of computing, but other times (as with most biological computing) the goal is just to recreate normal computation (or a subset of it) at a different scale or in more natural ways. Of course, these two approaches aren’t mutually exclusive! Imagine how great it would be if we could grow computers on the level of cells, or smaller. For starters, this approach could revolutionize health-care: you could program some of your own cells to sense and record your internal environment and release drugs only when necessary. It could also alter how we manufacture things; if you throught 3D printers are cool, what if you could program nanoscale assemblies?
 To start, it is important to understand what cells can already compute. For Luca Cardelli — first presentation on the second day of the 2nd workshop on Natural Algorithms and the Sciences — computation is the primary function of biology and so asking “what does the cell cycle switch computes” is perfectly natural. In an agent-based model of chemical reactions that you might see inside the membrane of a P-system, Cardelli & Csikász-Nagy (2012) showed that the cell cycle switch robustly implements the Angluin, Aspnes, and Eisenstat (2008) approximate majority algorithm from distributed computing. The goal of approximate majority is to figure out how to switch a cell from expressing some A and B to expressing only A or only B depending on which is in the majority of the initial configuration. Cardelli’s result is stunning since the approximate majority algorithm is optimal for a fault-tolerant implementation where agents are drawn uniformly at random to interact, and since the simplest network to implement it for an artificial cell system has the same structure as the empirically observed biological network. I would like to know if this sort of complex function could emerge in Valiant’s (2009) machine learning model of evolvability. Cardelli & Csikász-Nagy (2012) rely primarily on simulations to establish their results, but do not succumb to the curse of computing and present an analytic treatment.
To start, it is important to understand what cells can already compute. For Luca Cardelli — first presentation on the second day of the 2nd workshop on Natural Algorithms and the Sciences — computation is the primary function of biology and so asking “what does the cell cycle switch computes” is perfectly natural. In an agent-based model of chemical reactions that you might see inside the membrane of a P-system, Cardelli & Csikász-Nagy (2012) showed that the cell cycle switch robustly implements the Angluin, Aspnes, and Eisenstat (2008) approximate majority algorithm from distributed computing. The goal of approximate majority is to figure out how to switch a cell from expressing some A and B to expressing only A or only B depending on which is in the majority of the initial configuration. Cardelli’s result is stunning since the approximate majority algorithm is optimal for a fault-tolerant implementation where agents are drawn uniformly at random to interact, and since the simplest network to implement it for an artificial cell system has the same structure as the empirically observed biological network. I would like to know if this sort of complex function could emerge in Valiant’s (2009) machine learning model of evolvability. Cardelli & Csikász-Nagy (2012) rely primarily on simulations to establish their results, but do not succumb to the curse of computing and present an analytic treatment.
Moving to more complicated macromolecules, Jack Lutz — the 6th speaker on the 1st day — introduced us to Erik Winfree’s (1998) model of self-assembly by DNA tiles. In this model, we consider square tiles of DNA that have specific glue types and strength (0, 1, or 2) on their sides (corresponding to the pattern of A,C,G,T sticking out). The tiles are randomly deposited on a 2D surface and they remain attached if they can join up against other tiles with matching glue types. If the total glue strength is 2 or more (in other words, they must connect to a top and left tile of glue strength 1 or to a single other tile of strength 2) then the new tile remains attached, else it is stripped away by Brownian motion. This model is Turing Complete, and so can implement any computation, however this does not satisfactory. For Lutz, there is a more important concept of faithful simulation, where the tile system not only implements the same language, but also follows the same dynamics as the system it is simulating, including the same order of tiling. Under this more stringent definition of simulation, Doty et al. (2012) showed that there exists a DNA tiling systems can still simulate any other DNA tiling system. Thus, DNA tiling systems are programmable and controllable, but the challenge of experimental realization remains.
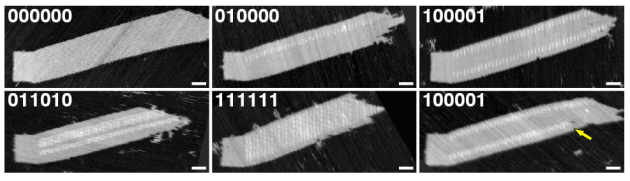
Rebecca Schulman — the 6th speaker on the 2nd day — focused more closely on experimental realization and applications in biology and engineering. In particular, she looked at home tiling systems can capture morphogenesis and the emergence of Turing patterns by modifying the tiling system for self-replication: the DNA crystal would be allowed to grow for a while and then get scissored into two parts that are then separated to continue growing on their own (Schulman, & Winfree, 2005). In Barish et al. (2009), she was able to experimentally work towards self-similarity and Lutz’ goal of basic programmability of the growing crystal by setting the initial seed. Challanges remain, and Schulman stressed in her presentation that although theoretical fault-tolerance results exist, the schemes are not currently implementable and the system remains non-resilient to noise and we know that can ruin universality. Still, it is amazing to see programmable DNA crystals growing at the scale of nanometers, even if we see mutations like the one highlighted by the yellow arrow in the figure below (Barish et al., 2005):
Note: this is the fifth blogpost of a series (1, 2, 3, and 4) on the recent workshop on Natural Algorithms and the Sciences at Princeton’s Center for Computational Intractability. There is one more post and a general overview still to come.
References
Angluin, D., Aspnes, J., & Eisenstat, D. (2008). A simple population protocol for fast robust approximate majority. Distributed Computing, 21(2), 87-102.
Barish, R. D., Schulman, R., Rothemund, P. W., & Winfree, E. (2009). An information-bearing seed for nucleating algorithmic self-assembly. Proceedings of the National Academy of Sciences, 106(15), 6054-6059.
Cardelli L, & Csikász-Nagy A (2012). The cell cycle switch computes approximate majority. Scientific Reports, 2 PMID: 22977731
Doty, D., Lutz, J. H., Patitz, M. J., Schweller, R. T., Summers, S. M., & Woods, D. (2012, October). The tile assembly model is intrinsically universal. In Foundations of Computer Science (FOCS), 2012 IEEE 53rd Annual Symposium on (pp. 302-310). IEEE.
Schulman, R., & Winfree, E. (2005). Self-replication and evolution of DNA crystals. In Advances in Artificial Life (pp. 734-743). Springer Berlin Heidelberg.
Valiant, L.G. (2009) Evolvability. Journal of the ACM 56(1): 3
Winfree, E. (1998). Algorithmic self-assembly of DNA. Doctoral dissertation, California Institute of Technology.

Pingback: Microscopic computing in cells and with self-as...
Reblogged this on Khuram Ali.
Pingback: Machine learning and prediction without understanding | Theory, Evolution, and Games Group
Pingback: Toward an algorithmic theory of biology | Theory, Evolution, and Games Group
Pingback: Programming language for chemistry | Theory, Evolution, and Games Group
Pingback: Cataloging a year of blogging: the algorithmic world | Theory, Evolution, and Games Group
Pingback: Cataloging a year of blogging: cancer and biology | Theory, Evolution, and Games Group
Pingback: EGT Reading Group 51 – 55 and a photo | Theory, Evolution, and Games Group
Pingback: The science and engineering of biological computation: from process to software to DNA-based neural networks | Theory, Evolution, and Games Group